Launch of a New Spike-In RNA Variant Module with 4 – 12 kb long RNAs
Today we launched the Long SIRV module. The long SIRVs are the latest addition to the Spike-In RNA Variants (SIRVs), Lexogen’s portfolio of external RNA reference standards. The 15 RNAs in this module cover the five length categories of 4 kb, 6 b, 8 kb, 10 kb, and 12 kb, each represented by three unique transcripts. These novel spike-in RNAs enable a thorough assessment of the representation of the length aspect in gene expression analyses.
By using these external standards, RNA-Seq pipelines running on Oxford Nanopore Technologies™ and Pacific Biosciences™ long-read platforms can finally be assessed for their performance in analyzing RNAs matching and exceeding the average mRNA length in eukaryotes. The currently available spike-in RNAs do not exceed 2.5 kb in length. Hence, only with the long SIRVs aspects such as processivity of reverse transcription and direct RNA-Seq, PCR length bias, full-length coverage, length restrictions during purifications, and performance of data analysis pipelines can be assessed and validated. On short-read platforms such as Illumina®, the handling of long RNAs by the respective library preparation methods and the re-assembly of these lengthy transcripts by the data analysis pipeline can be monitored.
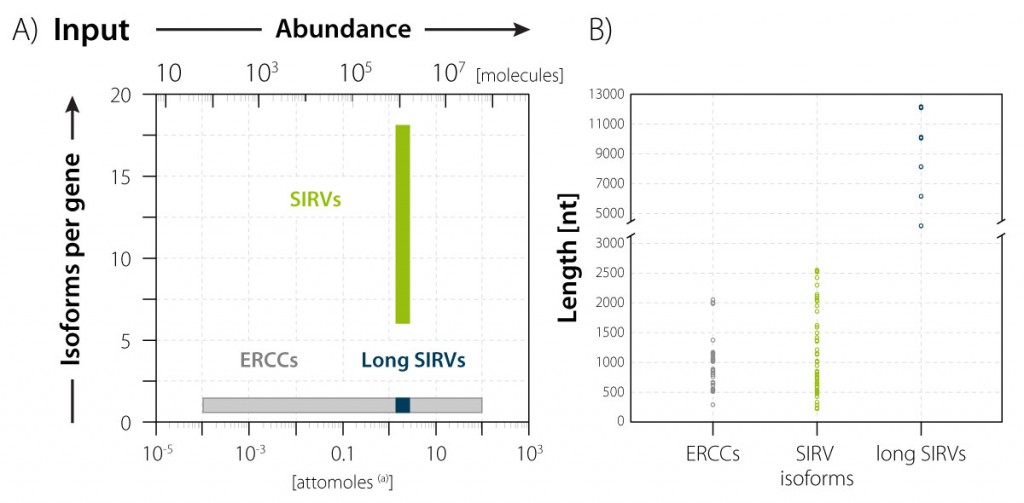
The long SIRV module is available in Lexogen’s SIRV-Set 4 in combination with two other modules, the SIRV Isoforms and the ERCC Mix. The 69 SIRV isoforms (mapping to 7 gene loci) comprehensively reflect splicing and transcriptional complexity, and the 92 ERCC transcripts cover the abundance aspect of transcriptome complexity by spanning a concentration range of 6 orders of magnitude (Fig. 1A). The long SIRVs and the SIRV Isoforms are present in equimolar concentrations in SIRV-Set 4, thereby clearly assigning the isoforms, concentration, length aspects of transcriptome complexity (Fig. 1B) to individual modules.
The sequences of all SIRV and ERCC transcripts do not overlap (in a relevant search window) with any transcript annotated in public data bases; NGS reads derived from spike-in RNAs can therefore be uniquely assigned to the “SIRVome”, the entirety of the annotated spike-in sequences (Fig. 2).
Usually, only 1% of all sequencing reads needs to be dedicated to the SIRVs for a comprehensive assessment, and since a tube with 10 µl SIRV-Set 4 is sufficient to spike 1000s of samples, all samples in an experiment can be routinely spiked to determine concordance of the resulting data sets.