New Globin Block Modules for QuantSeq Open Up an Opportunity for Cost-Efficient Gene Expression Analysis in Whole Blood Samples
We are happy to announce the release of the new Globin Block Modules for QuantSeq kits. These modules extend the functionality of the QuantSeq 3’ mRNA-Seq Library Prep, for gene expression profiling of whole blood, which is one of the most challenging sample types for transcriptome studies.
Blood is a highly informative, fairly low cost, and minimally invasive sample type widely used in clinical research. Furthermore, RNA sequencing of blood samples is a favored approach for gene expression aimed at biomarker discovery and investigating physiological and pathophysiological processes. However, the high abundance of globin RNA in whole blood, which consumes about 50-80% of sequencing reads, presents a huge challenge for analysis of transcripts at lower concentrations.
Lexogen’s new Globin Block Modules for QuantSeq allow cost-efficient reduction of globin mapping reads by up to 91%. Using these modules thousands of additional genes can be detected and quantified. Unlike other currently available globin depletion methods, which remove globin mRNAs from whole blood RNA prior to library preparation, the new modules enable globin reduction during library preparation itself.
The modules are designed in a way that the generation of cDNA library fragments from globin mRNAs is blocked, without adding any extra steps to the QuantSeq protocol. The modules consist of a single solution of Globin Blockers, which simply replaces the standard RNA Removal Solution from the QuantSeq kit (Figure 1).
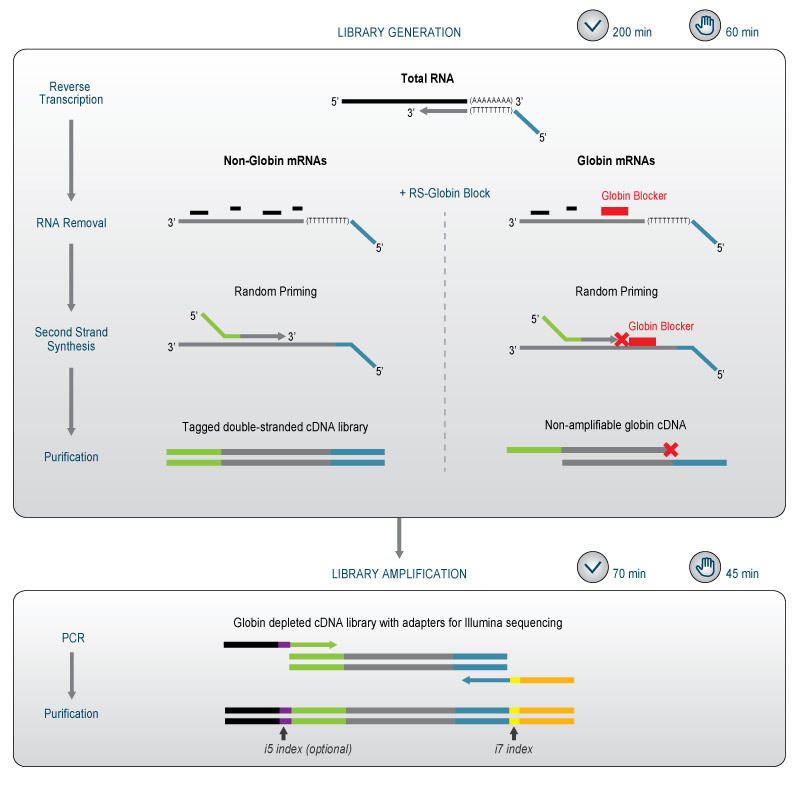
Figure 1. Schematic overview of the QuantSeq workflow using Globin Block Modules for globin depletion. Whole blood total RNA contains a mixture of both globin and non-globin mRNAs, which are both reverse transcribed by oligo(dT) priming during first strand synthesis. The RS-Globin Block (RS-GB ) solution is then added instead of the standard RNA Removal Solution (RS ). Following RNA removal, the Globin Blocker oligos bind specifically to globin first strand cDNA, downstream of random primers and block globin second strand cDNA synthesis. After second strand synthesis the sample will contain both non-amplifiable globin cDNA fragments and tagged double-stranded cDNA library fragments for non-globin mRNAs. During PCR, only the non-globin library fragments are amplified to add full-length adapters and indices for Illumina sequencing.
The Globin Block Modules are available for human (RS-Globin Block, Homo sapiens, Cat. No. 070.96) and pig (RS-Globin Block, Sus scrofa, Cat. No. 071.96) and contain enough volume for 96 reactions. The minimum input for QuantSeq library prep with Globin Block is only 50 ng of whole blood total RNA – 20 times lower than for other methods on the market.
QuantSeq 3’ mRNA-Seq Library Prep technology has already been used extensively for gene expression analysis in various sample types, e.g. tissues, cells, and FFPE samples. Thanks to its short workflow, low price, high sensitivity, and reproducibility it has become the method of choice in high throughput as well as smaller-sized projects.
The additional cost of using the Globin Block Modules together with QuantSeq adds only USD 4.98 per sample. This makes QuantSeq the most cost-efficient method available on the market for gene expression analysis in a wide range of sample types.
Learn more:
Visit the Globin Block Module page in our web store
Check the Globin Block Modules Instruction Manual
Visit the QuantSeq 3’ mRNA-Seq Library Prep web page
Contact us if you have any questions at [email protected].


